The translocator protein is a marker of inflammation in the brain, neuroinflammation, which is implicated in diseases such as multiple sclerosis and Alzheimer’s disease.
An understanding of the structure and function of this protein is vital in developing drugs to treat neuroinflammation. ANSTO’s research uses neutron reflectometry to study the structure of the translocator protein at the molecular level, to gain a deeper understanding of the protein and its interactions with potential drugs.
The translocator protein
The translocator protein (TSPO) is an integral membrane protein located primarily in the outer mitochondrial membrane of cells in a variety of peripheral tissue, and in immune cells in the brain [1]. It is highly conserved throughout evolution, indicating an important functional role for TSPO [1]. There is evidence that TSPO acts as a transport channel for cholesterol, a necessary step in steroid synthesis, and potentially acts as an ion channel [1].
Furthermore, TSPO has been implicated in inflammatory disease processes as it is greatly up-regulated in activated immune cells [2]. The synthetic TSPO ligand PK11195 has been shown to induce functional responses via TSPO, such as mitochondrial Ca2+ release, and potentially to modulate cholesterol transport and steroid production [1].
Despite the functional importance of TSPO, there is little structural data on the protein, due in large part to the difficulty of studying the structures of integral membrane proteins. The formation of a channel that can transport cholesterol was predicted using molecular dynamics [3], and a potential cholesterol recognition site was identified through nuclear magnetic resonance spectroscopy on a TSPO peptide [4].
However, further structural evidence of channel formation is needed, especially in relation to the effects of ligand binding on TSPO conformation, so that this can be correlated with the observed functional effects.
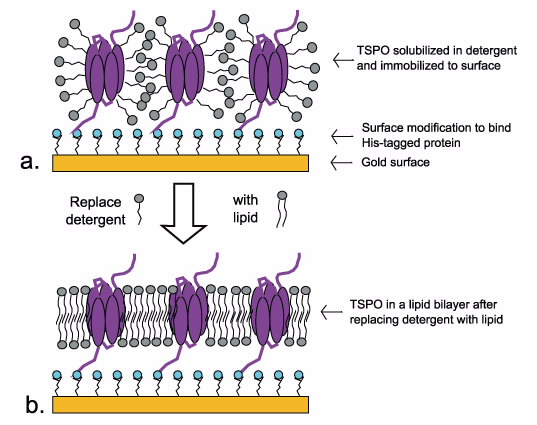 |
| Process of forming a protein- lipid bilayer. Detergent solubilised protein is immobilised to the surface (a), and detergent is gradually replaced with lipid. |
Using neutrons to study the TSPO
Neutron reflectometry makes it possible to study a membrane protein such as TSPO in a more natural lipid environment, which is extremely difficult using other structural techniques such as crystallography. The use of neutrons, in particular, allows contrast variation (the difference in scattering between hydrogen and deuterium) to be exploited.
Measurements can be made with either deuterated lipids or deuterated protein, and in multiple solvent contrasts to ‘match out’ certain components. This aids in structural refinement, and allows for the selective analysis of the different components of the system that cannot be distinguished using other methods.
We used neutron reflectometry in combination with our quartz crystal microbalance (Table 1) to study the formation of mixed TSPO – lipid bilayers, with a view to investigating changes associated with ligand binding.
Formation of TSPO – lipid bilayers
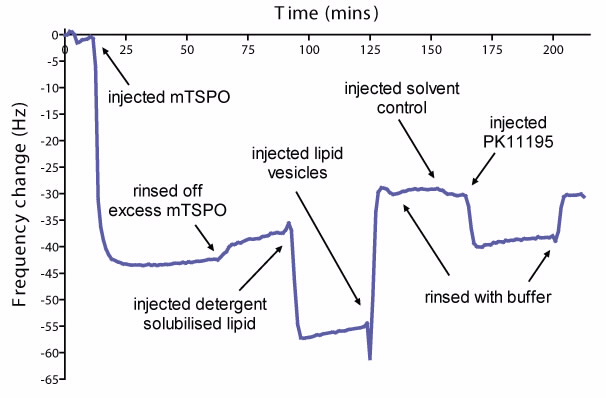 | 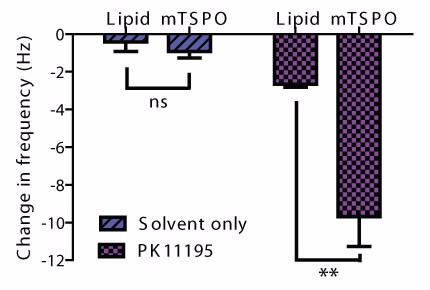 |
| (a) The change in frequency at each step of the formation of a TSPO-lipid bilayer, and interaction with the ligand PK11195. (b) Comparison of the change in frequency as a result of interaction with solvent only or Pk11195 on lipid bilayer with or without TSPO. Errors are standard deviations, n=4 in each group. |
We used the quartz crystal microbalance with dissipation monitoring (QCM-D) technique to monitor the formation of protein – lipid bilayers. First, we immobilised mouse TSPO (mTSPO) solubilised in detergent to a surface engineered to bind the protein (Fig. 1a). Next, we formed a lipid bilayer around the protein layer by replacing detergent with lipid (Fig. 1b).
The use of QCM-D allows us to monitor the adsorption of protein and lipid onto a resonating crystal surface. A decrease in frequency of the crystal indicates that mass has been added to the crystal surface. We observed a decrease in frequency when mTSPO binds to the crystal surface, indicating a protein surface coverage of approximately 75% (Fig. 2a).
The final change in frequency, after replacing detergent with lipid, is consistent with the remaining area being filled by lipid (Fig. 2a). We then tested the effect of the TSPO ligand PK11195 on the frequency response, in an attempt to detect ligand binding. We incubated PK11195 with the protein-lipid bilayer, which resulted in a significant change (p < 0.005) in frequency compared to the interaction with the surface or lipid alone (Fig. 2b).
The change was greater than expected for the mass of the ligand, indicating a possible conformational change of the protein, such as the opening of a channel and increased water content of the layer.
We proceeded to study this TSPO-lipid bilayer system with neutron reflectometry, with a view to detecting changes in the hydration of the protein-lipid layer as a result of ligand binding. Neutron reflectivity was measured at the major stages of TSPO-lipid bilayer formation (Fig. 3). There was indeed evidence of adsorption of a protein/detergent layer to the surface, with an approximate thickness of 50 Å.
In addition, there was a change in reflectivity after exchanging detergent for lipid, indicating the addition of lipid to the protein layer. We are currently refining a model to describe the TSPO-lipid layer, and the effect of ligands on the hydration of the layer.
| Technique | What does it measure? | What can we learn? |
| Quartz crystal microbalance with dissipation monitoring | The mass and rigidity of a film (such as a lipid bilayer) deposited on a surface. | We can follow the formation of a TSPO-lipid bilayer in real- time. We can learn whether the bilayer is rigid or "floppy", estimate the protein/ lipid coverage from the mass on the surface, and calculate the thickness of the layer. We can also follow changes in mass and rigidity during ligand binding.
|
| Neutron Reflectometry | The thickness and composition of films on a surface. | We can use the data to model the thickness and the composition of theTSPO-lipid bilayer. We can estimate the ratio of protein to lipid in the bilayer, and the thickness of the TSPO across the bilayer, giving us more information about the dimensions of the TSPO in the membrane environment. |
Outlook
Currently, we are working on improving the stability of the TSPO-lipid bilayer system for further studies with neutron reflectometry. The data collected on this system will make a valuable addition to the limited structural data available for the TSPO.
In particular, it could provide evidence for channel formation induced by ligand interactions, making it possible to relate changes in conformation to observed functional changes. This fundamental information could have impact in the broader context of understanding the function of TSPO and how it relates to inflammatory disease.
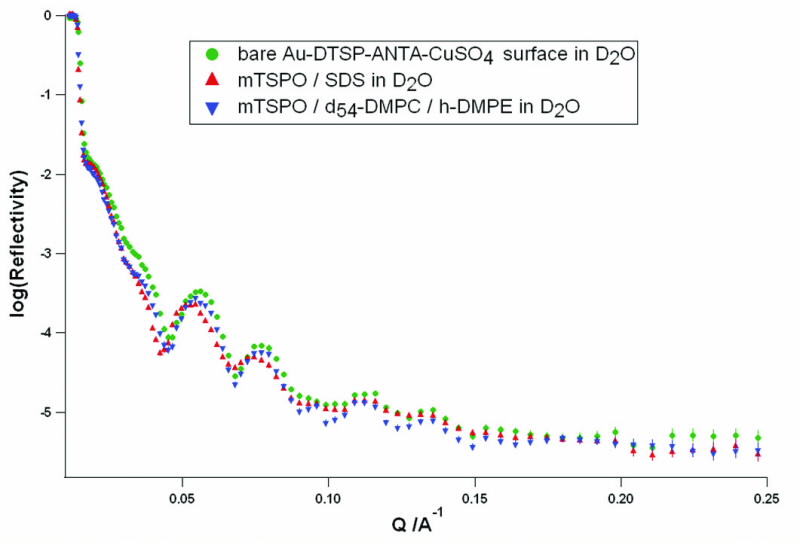 |
| Neutron reflectivity profiles at each stage of protein- lipid bilayer formation measured in D2O. Measurements were taken of the bare surface (green) the surface after addition of protein solubized in detergent (red), and after the replacement of detergent with lipid (blue). All measurements were made on the Surf reflectometer, ISIS UK. |
Authors
Claire Hatty1,2, Anton Le Brun1, Vanessa Lake1, Guo Jun Liu1,2, Richard Banati1,2
1ANSTO, 2University of Sydney
References
- Papadopoulos, V., Baraldi, M., Guilarte, T. R., Knudsen, T. B., Lacapere, J-J., Lindemann, P., Norenberg, M. D., Nutt, D., Weizman, A., Zhang, M-R., and Gavish, M., Translocator protein (18 kDa): new nomenclature for the peripheraltype benzodiazepine receptor based on its structure and molecular function, Trends in Pharmacological Sciences, 27(8), 402-409 (2006).
- Banati, R. B., Visualising microglial activation in vivo, Glia, 40(2), 206-217 (2002).
- Bernassau, J. M., Reversat, J. L., Ferrara, P., Caput, D., and Lefur, G., A 3D model of the peripheral benzodiazepine receptor and its implication in intra mitochondrial cholesterol transport, Journal of Molecular Graphics, 11, 236-244 (1993).
- Jamin, N., Neumann, J-M., Ostuni, M. A., Vu, T. K. M., Yao, Z-X., Murail, S., Robert, J-C., Giatzakis, C., Papadopoulos, V., and Lacapere, J-J., Characterization of the Cholesterol Recognition Amino Acid Concensus Sequence of the Peripheral-Type Benzodiazepine Receptor, Molecular Endocrinology, 19(3), 588-594 (2005).


