Self-assembled peptide systems, in particular those which form fibre networks and gels, may produce environments much like the extra-cellular matrix. Because they are easy to synthesize, biocompatible, and tuneable in structure there is a great scope for their use in drug delivery, cell culture and to promote the growth of neural cells.
For this reason the use of small-angle neutron scattering (SANS), and its ability to investigate the three-dimensional structural of networks in solution, is an ideal tool to investigate the effects of small chemical manipulations on the structure and kinetics of the assembly of gels into structures in situations approximating the extra cellular matrix.
In a recent publication in the open access journal Scientific Reports, researchers from the University of New South Wales use the SANS instrument QUOKKA to understand the assembly of two diphenylalanine-peptide-based hydrogels into structures, turning their attention to both the process of assembly and the structure of the final gel.
 |
| Figure 1: The left-hand side is the indole-based diphenylamine peptide and the right-hand side the carbazole-based peptide. |
Two different peptides were examined, both diphenylamine based, but with different end groups, one with an indole group and the other a carbazole group (Figure 1).
By observing and comparing the SANS over a period of 4 hours for the two gelators at high pH, it was shown that the indole peptide formed smaller less-well-defined fibres whereas the carbazole peptide formed a network of strongly associated fibres almost immediately.
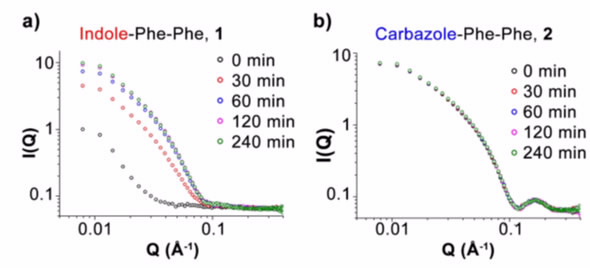 |
| Figure 2: Time evolution of SANS curves for the indole a), and carbazole b), diphenylalanine- based peptides show significant differences the time progression and final structures. |
The interpretation of SANS experimental results (Figure 2) were supplemented with real-space techniques (atomic-force microscopy and transmission electron microscopy) which gave a familiar representation of the structure as well examining the 1H NMR signal which is indicative of the monomeric population in the solution.
“Really it is the ability of SANS to probe simultaneously three-dimensional structure over a range of length-scales from the single nanometre to the 100’s of nanometers which made this insight possible” remarks Chris Garvey, the instrument scientist on QUOKKA and co-author on the paper.
Full reference: Martin, A. D., Wojciechowski, J. P., Robinson, A. B., Heu, C., Garvey, C. J., Ratcliffe, J., Waddington, L., J. Gardiner, J., Thordarson, P. (2017). Controlling self-assembly of diphenylalanine peptides at high pH using heterocyclic capping groups. Scientific Reports, 7, 43947. doi:10.1038/srep43947
Published: 10/03/2017


