 |
| Fig 1- Cartoon of a flagellum - the FliG ring is part of the aquamarine double-disc structure in the lower membrane |
A paper just published in Nature Structural and Molecular Biology, involving Institute staff, and data taken on the Small-Angle X-ray Scattering beamline at the Australian Synchrotron, demonstrates that Escherichia coli FliG, one of the earliest-assembling flagellar motor proteins, forms ordered ring structures via domain-swap polymerization.
Solution structural data, in combination with in vivo biochemical cross-linking experiments and evolutionary covariance analysis, reveals that FliG exists predominantly as a monomer in solution but only as domain-swapped polymers in assembled flagellar motors. A general structural and thermodynamic model for self-assembly is proposed, in which a structural template controls assembly and shapes polymer formation into rings.
The work is a collaboration involving the Victor Chang Cardiac Research Institute, European Molecular Biology Laboratory Australia, Oxford and Osaka Universities and ANSTO.
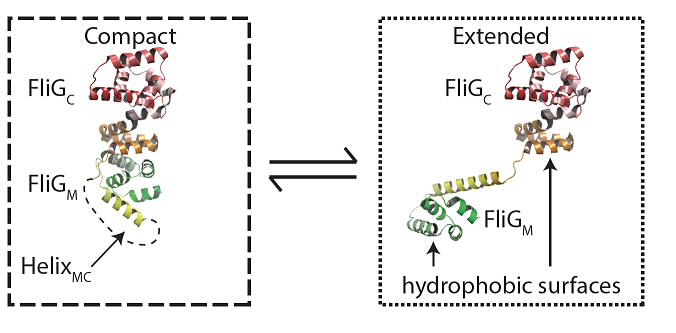 |
| Fig 2- The structure transition studied in the paper |
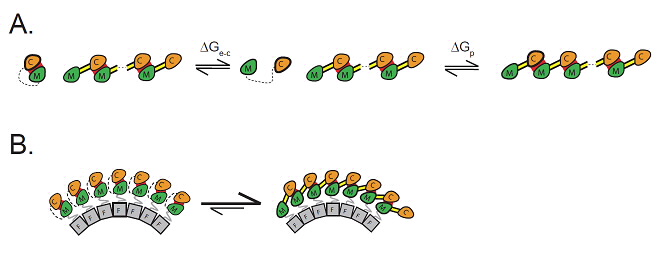 |
| Fig 3- Cartoon illustrating the proposed mechanism of ring formation. |
The full reference is M. A. B. Baker, R. M. G. Hynson, L. A. Ganuelas, N. S. Mohammadi, C. W. Liew, A. A. Rey, A. P Duff, A. E Whitten, C. M. Jeffries, N. J. Delalez, Y. V. Morimoto, D. Stock, J. P. Armitage, A. J. Turberfield, K. Namba, R. M. Berry and L. K. Lee, "Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor", Nature Structural and Molecular Biology doi:10.1038/nsmb.3172 (2106).
Published: 18/02/2016

