Researchers have developed models of bacterial outer membranes that can help develop better antibiotics to fight antibiotic-resistant bacteria.
 |
| An Artist's Impression (David S. Goodsell) of molecules in a slice of E.coli cell. The green areas show the membrane under study |
Introduction
Biological membranes are watertight barriers that regulate what gets in and out of cells. Some membranes have very specialised functions, such as insulation for nerve cells or capturing light in the rod cells of the eye.
Currently, around the world, there are significant research efforts in developing antibiotics that can attack bacterial membranes to help combat the threat posed by antibiotic-resistant bacteria.
As membranes are highly complex structures models, or man-made copies of a bacterial membrane, are needed to better understand the effectiveness of a potential new drug.
In this study we have developed models of bacterial outer membranes and investigated their molecular structure using neutron reflectometry. The results show that the models we are creating of the bacterial outer membrane are biologically relevant, reproducible and can help test and develop better antibiotics.
All cells, whether bacterial or from higher organisms are surrounded by a membrane. The membrane is what separates the cell from its external environment. The membrane is made of a lipid bilayer which acts as a hydrophobic (water resistant) barrier. Distributed within the membrane are membrane proteins, which act as the cell’s gate keepers, allowing nutrients into the cell and letting the waste products out.
The importance of cell membranes is also underlined by the fact that over 60% of currently available drugs and 40% of new drugs target membrane proteins [1] and there is a large focus on antibiotics that attack the lipid-bilayer component of bacterial membranes.
To better understand a membrane we need to understand the physical properties of the lipid bilayer and how membrane proteins function and assemble.
This is not a trivial task. A cell membrane is a very complex structure with many different types of lipids and an even greater number of different proteins, all contributing to the function of the cell.
To study biological membranes we take a reductionist approach i.e. we start with a very simple model, see how that works and then slowly build up in complexity. So our research seeks to investigate how to make these simple models and whether they mimic what is seen in nature.
So why use neutrons?
There are a number of reasons why scientists studying membranes want to use the neutrons produced by the OPAL reactor at ANSTO.
The main neutron technique is neutron reflectometry, which provides a depth profile of the 5 to 6 nanometre thick membrane over a large area, whereas techniques such as microscopy only allow study of the initial membrane surface over small areas.
Most proteins in the cell are surrounded by water. However, because membrane proteins reside in the water-resistant phospholipid bilayer this makes them difficult to study using many of the routine scientific techniques such as X-ray crystallography.
Neutron reflectometry provides structural information on membrane proteins in their natural bilayer surrounding, by taking a slice through the depth of the membrane.
Reflectometry tells us the thickness and distribution of the different components (typically lipids, proteins and water) as we measure through the depth of the bilayer.
Characterising model bacterial outer membranes
Bacteria that have a double membrane are classed as Gram-negative. Gram-negative bacteria include bacteria such as E. coli and Salmonella.
They have an inner membrane and an outer membrane. The outer membrane is unusual in that it is asymmetric. An asymmetrical membrane is one in which the half
of the membrane that faces the inside of the cell is very different from the half that faces the outside of the cell. We have made model membranes that replicate this asymmetry, using floating supported bilayers [2].
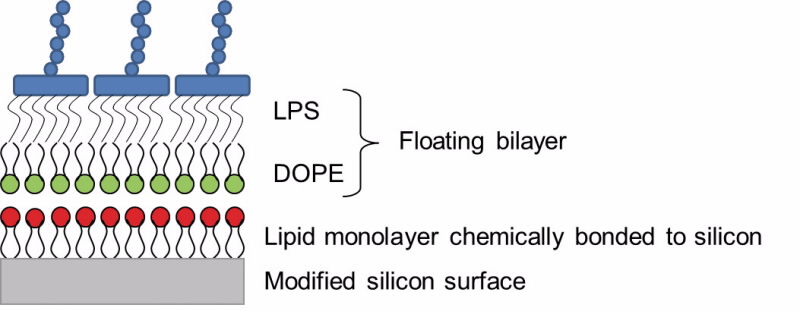 |
| Fig 1. A schematic showing the principle of a floating supported bilayer. A lipid monolayer is chemically bonded to a modified silicon surface. Then using Langmuir-Blodgett deposition the lower lipid leaflet is deposited (in this case the DOPE layer) and then using Langmuir-Schaefer deposition an upper leaflet of lipids is deposited (the LPS layer). |
Floating supported bilayers are advantageous as the membrane is not physically bonded to the surface that supports it. This means that the properties of the model membrane are not influenced by the supporting surface and should behave more like a membrane found in nature.
A silicon surface is used both because the surface can be made atomically flat, which is necessary for good neutron reflectometry measurements, and also because silicon is amenable to a number of different chemistries, making the surface easy to functionalise.
To create the floating supported bilayer, a lipid monolayer is grafted onto a silicon surface. The bilayer that we wish to create then sits or floats above the grafted monolayer (Fig. 1). The membrane is created by using a series of Langmuir-Blodgett and Langmuir-Schaefer dipping techniques [3].
Using a state-of-the-art Langmuir dipping trough, the modified silicon surface is pushed through a monolayer of lipid on a liquid surface either vertically (Langmuir-Blodgett) or horizontally (LangmuirSchaefer). When the silicon surface is passed through the lipid monolayer the material transfers from the liquid surface onto the silicon creating the floating bilayer.
Using the dipping techniques we have managed to create for the first time asymmetric floating bilayers of di-oleoyl-phosphatidylethanolamine (DOPE) and lipopolysaccharide (LPS). DOPE is the most abundant lipid on the inner side of the membrane of the outer membrane, whilst LPS is the abundant molecule on the outer side of the outer membrane.
These floating bilayers have been found to be good models, as they can replicate asymmetry, mimic the phase behaviour of lipids in nature and mimic the fluidity of bilayers. Using neutron reflectometry on the Platypus reflectometer at OPAL we have been able to characterise the structures of these model bacterial outer membranes. Fig. 2 shows the data collected on the reflectometer and the structure of the bilayer as a scattering-length, density profile.
A neutron scattering-length density (SLD) can be considered as the “neutron refractive index” and we model the change in scattering-length density through each layer of the membrane to deduce its structure (Fig. 2).
Using deuteration to get further details
Neutron scattering in soft matter and biomolecular science can be made more advantageous if some components of a multi-component system are deuterated. Neutrons scatter from hydrogen very differently than from its heavier isotope deuterium.
Therefore, if we label one of the components in the membrane system with deuterium, each component can be picked out of the complex system.
Deuterated LPS was made in the bio-labs of the National Deuteration Facility at ANSTO. The deuterated LPS allows us to distinguish between it and the hydrogenous DOPE leaflet when we swap between a D2O solvent and regular H2O, adding further detail to our model, such as the surface coverage and molecular area of each component in the membrane.
Summary
We have created robust models of the outer membrane of Gram-negative bacteria. Using neutron reflectometry and molecular deuteration we have been able to characterise these model membranes to see if they reflect the properties of a membrane in nature.
These model membranes are powerful tools for studying how membrane proteins function and how antibacterial agents and other drugs interact with membranes.
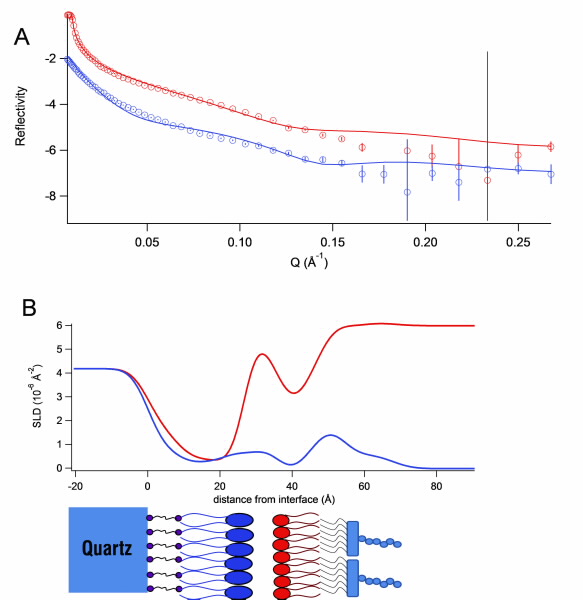 |
| Fig 2. A) The reflectivity data (points with error bars) and model fits (lines) of a floating model membrane of DOPE and lipopolysaccharide under a D2O (red) and H2O (blue) solvent contrast. B) The corresponding neutron scattering-length density profile. The schematic below the profile shows the position of each component along the membrane. |
Authors
Tomas Mayfield, Anton Le Brun, Stephen Holt
ANSTO
References
- Yildirim, M. A., Goh, K.-I., Cusick, M. E., Barabasi, A.-L., and Vidal, M., Drugtarget Network, Nature Biotechnology, 25(10), 1119-1126 (2007).
- Hughes, A. V., Howse, J. R., Dabkowska A., Jones, R. A. L., Lawrence M. J., Roser, S. J., Floating lipid bilayers deposited on chemically grafted phosphatidylcholine surfaces, Langmuir, 24(5), 1989-1999 (2008).
- Charitat, T., Bellet-Amalric, E., Fragneto, G., and Graner, F., Absorbed and free lipid bilayers at the solid-liquid interface. European Physical Journal B, 8(4), 583-593 (1999).


